-
Notifications
You must be signed in to change notification settings - Fork 3
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
M and N parameters #5
Comments
|
Thanks for your interest!
TLDRIf you want to replicate SMA using convolution with a kernel, the Long explanationConsider that the SMA method is based on creating a template by averaging and subsequent subtraction. The implementation in ArtACS is using convolution with a (comb) kernel. Therefore mathematics differ slightly, although results would be identical under most circumstances.
If we would use convolution, we would first have to define a kernel that does something similar to this. For this kernel, almost all values of the kernel are zeros. The width of the kernel defines how many periods (i.e. segments) one would "average" relative to the current sample. In that regard, the parameter You can find more details in the pdf accompanying the package: https://github.com/agricolab/ArtACS_preprint/blob/master/docs/main.pdf As a side note, consider that his package was written in 2017, but Kohli and Casson had published several papers describing their results for SMA on tACS artifacts already, e.g. Kohli 2015. |
|
Dear, |

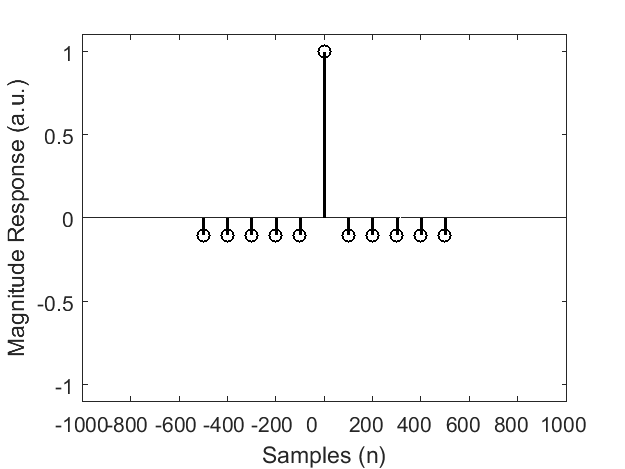
The comb filter is an implementation of the SMA method (Kohli 2019) if I am not wrong.
In that paper they have 2 parameters, M (neighboring segments) and N (the total number of segmented epochs).
In your implementation, what are the values of these 2 parameters?
Thanks
The text was updated successfully, but these errors were encountered: