Python/PyTorch source code for the paper:
A. Genovese,
"ALLNet: Acute Lymphoblastic Leukemia Detection using lightweight convolutional networks",
in Proc. of the 2022 IEEE Int. Conf. on Computational Intelligence and Virtual Environments
for Measurement Systems and Applications (CIVEMSA 2022),
Chemnitz, Germany, June 15-17, 2022, pp. 1-6
ISBN: 978-1-6654-3445-4. [DOI: 10.1109/CIVEMSA53371.2022.9853691]
Paper:
https://ieeexplore.ieee.org/document/9853691
Project page:
https://iebil.di.unimi.it/cnnALL/index.htm
Citation:
@InProceedings{civemsa22_all,
author = {A. Genovese},
booktitle = {Proc. of the 2022 IEEE Int. Conf. on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA 2022)},
title = {ALLNet: Acute Lymphoblastic Leukemia Detection using lightweight convolutional networks},
address = {Chemnitz, Germany},
pages = {1-6},
month = {June},
day = {15-17},
year = {2022},
note = {Accepted},}
Main files:
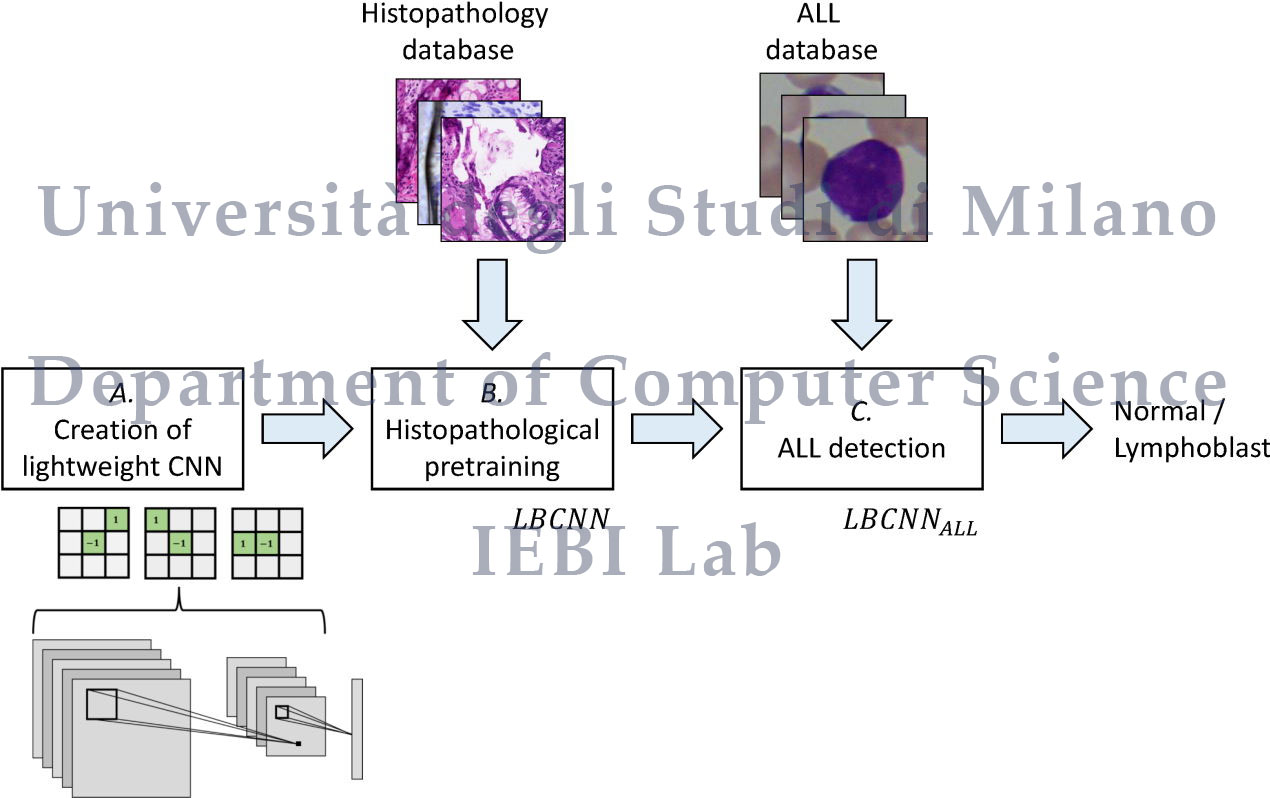
- 0_PyTorch_ADP_HistoNet_LBCNN/pytorch_adp_histonet.py: pretraining on ADP database;
- 1_PyTorch_ADP_HistoNet_LBCNN_fineTune_ALL_IDB/pytorch_adp_histonet_finetune_all.py: fine tuning on the ALL database.
Instructions:
-
cd to "0_PyTorch_ADP_HistoNet_LBCNN" and run "pytorch_adp_histonet.py" to pretrain the LBCNN on the ADP database, implemented according to the paper:
Required files:
-
ADP/img_res_1um_bicubic/
ADP database, split in patches, obtained following the instructions at:
https://www.dsp.utoronto.ca/projects/ADP/ -
ADP/ADP_EncodedLabels_Release1_Flat.csv file containing the labels of the ADP database, obtained following the instructions at:
https://www.dsp.utoronto.ca/projects/ADP/
-
-
Copy the trained models in "1_PyTorch_ADP_HistoNet_LBCNN_fineTune_ALL_IDB\pretrained_nets". For simplicity, some trained models are already present.
-
cd to "1_PyTorch_ADP_HistoNet_LBCNN_fineTune_ALL_IDB" and run "pytorch_adp_histonet_finetune_all.py" to train the ALLNet on the ALL-IDB database for Acute Lymphoblastic Leukemia detection.
Required files:
- /ALL_IDB2
ALL-IDB database, obtained following the instructions at: https://homes.di.unimi.it/scotti/all/
- /ALL_IDB2
The databases used in the paper can be obtained at:
-
Atlas of Digital Pathology (ADP)
https://www.dsp.utoronto.ca/projects/ADP/Mahdi S. Hosseini, Lyndon Chan, Gabriel Tse, Michael Tang, Jun Deng, Sajad Norouzi, Corwyn Rowsell, Konstantinos N. Plataniotis, Savvas Damaskinos "Atlas of Digital Pathology: A Generalized Hierarchical Histological Tissue Type-Annotated Database for Deep Learning" Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2019, pp. 11747-11756
-
Acute Lymphoblastic Leukemia Image Database for Image Processing (ALL-IDB)
https://homes.di.unimi.it/scotti/all/R. Donida Labati, V. Piuri, F. Scotti "ALL-IDB: the acute lymphoblastic leukemia image database for image processing" in Proc. of the 2011 IEEE Int. Conf. on Image Processing (ICIP 2011), Brussels, Belgium, pp. 2045-2048, September 11-14, 2011. ISBN: 978-1-4577-1302-6. [DOI: 10.1109/ICIP.2011.6115881]