-
Notifications
You must be signed in to change notification settings - Fork 3
Classification schema
One of the analysis steps performed by ProteinClusterQuant is the analysis of all protein pairs in the constructed protein-peptide network.
A protein pair is formed by two protein nodes sharing a peptide node, and that may have other peptide nodes unique to each one of them.
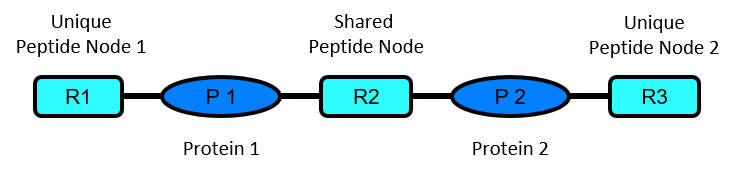
This analysis consists on the systematic detection of peptide node ratio outliers in each one of the protein pairs analyzed and classifies the protein pairs in different cases as described below. This classification schema is able to handle peptide nodes having infinity ratio values (only detected in one of the samples), but do not further consider protein pairs with peptide nodes that lack a quantitative measurement.
It further classifies all protein pairs according to the presence of peptides nodes with a significantly different ratio value, either being the shared peptide node or one (or both) of the unique peptide nodes. Alternatively, none of the peptide nodes may differ significantly.
In case a significantly different peptide node is present, protein pairs are further distinguished based on the peptide node’s connectivity: one edge (unique) or two edges (shared) or three or more edges (shared by a third or more protein nodes). A connectivity of more than two edges can explain the shared peptide node’s significance within the protein pair analyzed. If only two edges are identified, a significant ratio measurement of the shared peptide node remains unexplained. In this case the significance is either a result of an incorrect measurement or caused by an additional protein edge not identified in the experiment and thus remains unexplained.
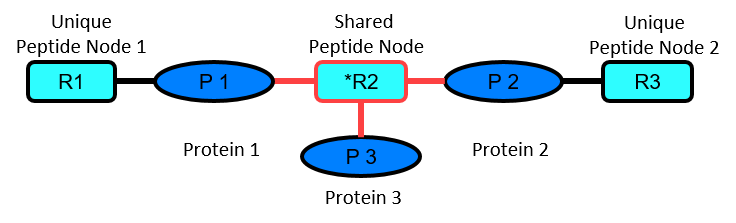
When analyzing protein pairs, this classification schema is used:
It is based on a user-defined cutoff threshold which is applied to the differences between the peptide node ratios. Depending on which peptide nodes are significantly different, the protein pair is classified accordingly. Optionally, if input parameter statisticalTestForProteinPairApplied is TRUE, a Iglewicz-Hoaglin statistical test is applied in order to consider that a unique peptide is an outlier or not.
| Classification code | Description |
|---|---|
| 1 | Shared peptide node is significantly different and is not shared by other protein node. |
| 2 | Shared peptide node is significantly different and is shared by other protein node. |
| 3 | Unique peptide node is significantly different, or the difference between the two unique peptide nodes is larger than threshold. |
| 4 | Both unique peptide nodes are significantly different and shared peptide node is significantly different in between. |
| 5 | There is not a significant difference. Difference between unique peptide nodes is less than threshold. |
| 6 | Not classified. There is not enough data, i.e. some of the unique peptide nodes is missing. |
Contact person:
Salvador Martínez-Bartolomé (salvador at scripps.edu)
Research Associate
The Scripps Research Institute
10550 North Torrey Pines Road
La Jolla, CA 92037
Git-Hub profile