-
Notifications
You must be signed in to change notification settings - Fork 3
XGMML files
XGMML files (the eXtensible Graph Markup and Modeling Language) are files that can be opened in Cytoscape. ProteinclusterQuant exports the data in that format in order to visualize the protein-peptide network and the quantitative analysis.
Each XGMML file contain a protein-peptide network, that can contain more than one protein cluster or connected component.
In order to open the network in Cytoscape:
1. Open Cytoscape.
2. Click on Import Network From File.

or go to menu: File -> Import -> Network -> File...
or press CTRL+L
3. Select the XGMML file you want to open. Then, the file will be loaded.
4. Click on Apply Preferred Layout in order to see the network properly in the screen.

Once you open the file, you can zoom in and zoom out to look for particular clusters and nodes.
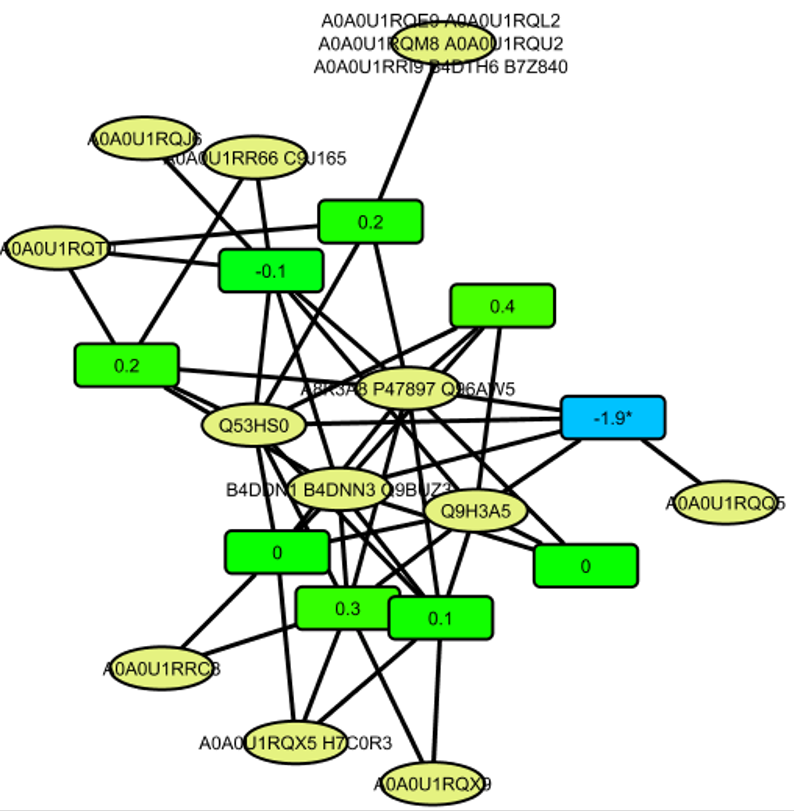
You can also use the Search text box on the top right in order to search for particular protein, peptides or any associated feature.
The following table shows which are the features that are encoded in the network and therefore are searchable through the Cytoscape Search feature:
| Feature name | Peptide Node | Protein Node | Description |
|---|---|---|---|
| ACC | UniProt protein Accessions | UniProt accession number, i.e. A2BC19, P12345, A0A022YWF9. If the protein node contains more than one protein, they will be listed separated by spaces. | |
| ID | Uniprot entry name | UniProt entry name (formerly called ID) associated with the protein, i.e. INS_HUMAN, INS1_MOUSE and INS2_MOUSE | |
| Classification1Case | 0-7 | See [[Classification Schemas | |
| Classification2Case | 1-7 | See [[Classification Schemas | |
| conclusiveProtein | Boolean | Whether the protein is linked to a unique peptide node | |
| conclusiveProteinNode | Boolean | Whether the protein node is linked to a unique peptide node | |
| FDR | Decimal | Quantification False Discovery Rate associated with the peptide node ratio. | |
| finalRatio | Decimal | Final ratio of the peptide node, log2 transformed | |
| GeneName | Gene ID | Gene ID associated with the protein. If the protein node contains more than one gene ID, they will be listed separated by scaces. | |
| lightIons | Integer | Number of Light ions associated with the peptide node (just in case of isobaric-isotopologue quantitation). | |
| heavyIons | Integer | Number of heavy ions associated with the peptide node (just in case of isobaric-isotopologue quantitation). | |
| ionCount | Integer | Number of ions associated with the peptide node (just in case of isobaric-isotopologue quantitation). It is equals to the sum of heavyIons + lightIons. | |
| isProtein | 0 | 1 | 0 for peptide nodes, and 1 for protein nodes |
| name | Peptide sequences | Protein accessions | Node name |
| numConnectedPeptideNodes | Integer | Number of connected peptide nodes associated with the protein node. | |
| numConnectedProteinNodes | Integer | Number of connected protein nodes associated with the peptide node. | |
| numMSRuns | Integer | Number of different MS runs in which the peptide node has been detected | |
| numPeptideSequences | Integer | Number of different peptide sequences included in the peptide node. | |
| numPeptideSequencesInProteins | Integer | Number of different peptide sequences connected to the protein nodes. | |
| numProteins | Integer | Integer | Number of proteins in the protein node, or number of different proteins connected to the peptide node |
| numPsms | Integer | Number of PSMs in the peptide node. | |
| numPsmsInProtein | Integer | Number of PSMs associated with the protein node | |
| PCQ_ID | Peptide sequences | Unique identifier for the peptide nodes containing the peptide sequences of the peptide node. | |
| PeptideSequences | Peptide sequences | Peptide sequence of the peptide node. It is equals to PCQ_ID. | |
| ProteinDescription | Protein description | Description of the proteins of the protein node. If the protein node contains more than one protein, the descriptions will be separated by '####' | |
| shared name | Peptide sequences | UniProt protein Accessions | Identifier of the node. |
| significant | Boolean | 0 for False, 1 for True | |
| Species | Species | Taxonomy species of the protein in the protein node. | |
| UniprotKB | UniProt protein Accessions | UniProt accession number. If the protein node contains more than one protein, they will be listed separated by spaces. | |
| uniquePeptide | Boolean | Whether the peptide is unique (is connected only to one protein) or not | |
| uniquePeptideNode | Boolean | Whether the peptide node is unique (is connected only to one protein node) or not | |
| Variance | Decimal | Calculated variance associated with the peptide node ratio. A lower variance means a more reliable ratio. | |
| Weight | Decimal | Calculated weight associated with the peptide node ratio. A higher weight means a more reliable ratio. |
Additionally, you can include new columns for annotations from UniprotKB with the option 'uniprot_xpath' in the input parameters file. Those annotations will be then searchable in Cytoscape. See input parameters file for more information.
Contact person:
Salvador Martínez-Bartolomé (salvador at scripps.edu)
Research Associate
The Scripps Research Institute
10550 North Torrey Pines Road
La Jolla, CA 92037
Git-Hub profile